Design multi state and functional proteins using sequence space diffusion model
November 6, 2024
Source: drugdu
 321
321
Protein Denoising Diffusion Probabilistic Models (DDPM) can efficiently generate novel proteins that meet specific properties and functional requirements, and have significant value in the field of protein design. Although many current models, such as RFdiffusion and Chroma, have shown excellent performance in generating protein three-dimensional structures, there are still challenges in generating proteins with sequence specificity and functional properties, and these models often require a lot of time and computational resources for training.
David Baker's team at the University of Washington has proposed a new solution to address this challenge. They believe that diffusion in sequence space can more effectively learn more knowledge from the current large amount of protein sequence data. Therefore, the research team developed a sequence space diffusion model ProteinGenerator based on RoseTTAFold, which can generate protein sequences and structures that meet the required properties. Subsequently, the research team further successfully designed multiple proteins with different functions using this model. In September 2024, the research findings were published in the journal Nature Biotechnology under the title "Multistate and Functional Protein Design Using RoseTTAFold Sequence Space Diffusion".
 Model Overview
Model Overview
The model architecture of ProteinGenerator is shown in Figure 1, which maps protein sequences to a high-dimensional continuous sequence space in the form of one hot codes and continuously diffuses them. In the process of trajectory inference, ProteinGenerator predicts the three-dimensional structure of the sequence through RoseTTAFold, and simultaneously mixes the embedding of noise sequence (Xt) and protein structure constraint (Yc), and obtains the protein sequence and three-dimensional structure through multiple denoising processes. The protein sequence will also be mapped back to the natural protein space through argmax operation to obtain the final protein sequence.
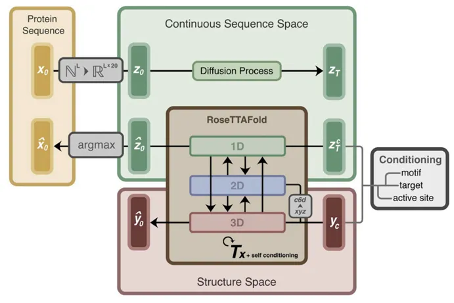 Figure 1. Schematic diagram of the model architecture of ProteinGenerator. Protein sequences will be mapped to high-dimensional space, and in the inference stage, protein sequences and corresponding structures will be predicted based on sequence and structural conditions.
Figure 1. Schematic diagram of the model architecture of ProteinGenerator. Protein sequences will be mapped to high-dimensional space, and in the inference stage, protein sequences and corresponding structures will be predicted based on sequence and structural conditions.
The inference trajectory of ProteinGenerator is shown in Figure 2. During each inference process, ProteinGenerator infers based on sequence guidance such as net charge, protein hydrophobicity, activity, and amino acid composition, as well as structural constraints such as secondary structure and three-dimensional coordinates. After multiple iterations, the protein sequence and structure that meet the expected requirements are finally obtained.
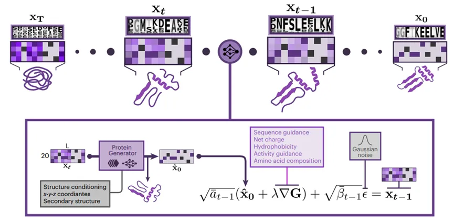 Figure 2. Schematic diagram of the inference trajectory of ProteinGenerator. At each step of the diffusion process, sequence X0 is generated by combining sequence Xt with structural information, sequence bias, and noise to produce the sequence and structure at time Xt-1. This process is repeated T times to obtain.
Figure 2. Schematic diagram of the inference trajectory of ProteinGenerator. At each step of the diffusion process, sequence X0 is generated by combining sequence Xt with structural information, sequence bias, and noise to produce the sequence and structure at time Xt-1. This process is repeated T times to obtain.
training method
ProteinGenerator uses sequence structure data from PDB database for training. The training process involves uniformly sampling time steps t within the interval [0, T], and gradually adding noise to the original sequence X0 until a sequence XT composed entirely of Gaussian noise is obtained. The main objective of the model is to predict the denoised sequence X0 and its corresponding structure. In terms of setting the loss function, researchers used classification cross entropy to evaluate the accuracy of sequence prediction, and introduced various structural loss calculation methods including FAPE, bond length, bond angle, etc. to evaluate the performance of structural prediction. In addition, the model also adopts a self conditioning mechanism, allowing prediction based on the prediction results of the previous time step and the backward calculated Xt-1 as conditions during the training and inference stages. In order to improve the generalization ability and robustness of the model, researchers also adopted a multi task learning strategy, combining standard diffusion tasks with structural prediction tasks and fixed skeleton sequence design tasks. This strategy helps ensure that the model can maintain consistency between sequence and structure during the diffusion process, thereby improving the overall prediction accuracy.
Application of ProteinGenerator in Protein Design
Design of proteins rich in rare amino acids
In order to evaluate the ability of ProteinGenerator to perform sequence structure inference outside of the training data distribution, researchers hope to design proteins rich in amino acids with structural or functional characteristics that have been undersampled during evolution. Therefore, researchers generated proteins containing high-frequency rare amino acids (tryptophan, cysteine, valine, histidine, methionine) through ProteinGenerator, and the sequences of these proteins were significantly different from those of natural proteins. The researchers subsequently screened the generated sequences and selected 96 sequences for experimental characterization while ensuring high confidence in the results predicted by AlphaFold2. As shown in Figure 3, ProteinGenerator can go beyond the composition of natural protein like sequences, infer sequence structure relationships, and design folded and thermally stable proteins with desired sequence characteristics.
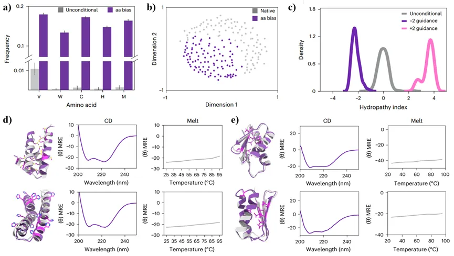 Figure 3. Design of proteins rich in rare amino acids. (a) (b) The frequency and spatial distribution of rare amino acids in unconditional sequences and rare amino acid sequences; (c) The hydrophobicity distribution of sequences generated based on hydrophobicity requirements and unconditional sequences; (d) (e) Circular dichroism (CD) and melting point test results of rare amino acid sequences, where gray and purple represent the structures generated by ProteinGenerator and the predicted results by AlphaFold2, respectively.
Figure 3. Design of proteins rich in rare amino acids. (a) (b) The frequency and spatial distribution of rare amino acids in unconditional sequences and rare amino acid sequences; (c) The hydrophobicity distribution of sequences generated based on hydrophobicity requirements and unconditional sequences; (d) (e) Circular dichroism (CD) and melting point test results of rare amino acid sequences, where gray and purple represent the structures generated by ProteinGenerator and the predicted results by AlphaFold2, respectively.
Design of repetitive proteins
Repetitive proteins contain a large number of tandem copies of sequence structural units, which play important roles in molecular recognition and signal transduction. The design of such proteins usually requires pre specification of structural features or calculation through Markov Monte Carlo, which often consumes a lot of time and computational resources. Fine tuning the ProteinGenerator by applying repetitive symmetry to the noise sequence distribution at each time step and constraining it with specified secondary structures can quickly generate repetitive proteins. Researchers characterized 74 capped repetitive proteins and 86 uncapped repetitive proteins generated by ProteinGenerator through experiments. Among them, 27 capped repeat proteins and 10 uncapped repeat proteins can be detected by volume exclusion chromatography (SEC) and are soluble monomers. The circular dichroism results indicate that 7 out of the 8 proteins tested have the expected secondary structure.
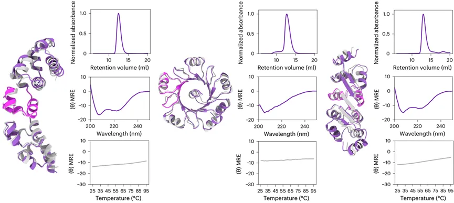 Figure 4. Design of repetitive proteins. The gray structure and purple structure represent the structures predicted by ProteinGenerator and AlphaFold2, respectively, while pink represents the asymmetric parts of the structure.
Figure 4. Design of repetitive proteins. The gray structure and purple structure represent the structures predicted by ProteinGenerator and AlphaFold2, respectively, while pink represents the asymmetric parts of the structure.
Design of Bioactive Peptide Cage
The design of proteins that activate functions based on environmental conditions is very attractive for fields such as drug design. Therefore, researchers further utilized ProteinGenerator to design an active peptide cage containing bee venom peptides. After cleavage by Flynn protease, the active peptide cage releases the bee venom peptides inside. For ProteinGenerator, it is only necessary to specify sequences and conditions with specific functions, and allow other sequences to diffuse freely to generate active peptide cages that release proteins with expected functions under specific conditions.
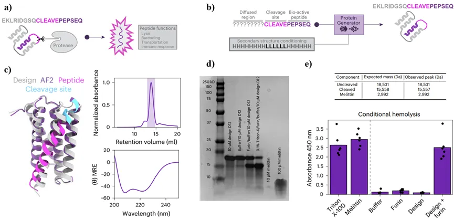 Figure 5. Design of an active peptide cage containing bee venom peptide. (a) (b) The principle of generating active peptide cages using ProteinGenerator; (c) The structure of the active peptide cage, where pink represents peptide segment D12 used for downstream analysis; (d) (e) Validation of peptide cage related peptide segments before and after cleavage.
Figure 5. Design of an active peptide cage containing bee venom peptide. (a) (b) The principle of generating active peptide cages using ProteinGenerator; (c) The structure of the active peptide cage, where pink represents peptide segment D12 used for downstream analysis; (d) (e) Validation of peptide cage related peptide segments before and after cleavage.
Design of multi state proteins
The researchers also explored the potential of ProteinGenerator in generating different conformations of the same protein sequence. In order to meet the task objectives, the researchers inputted the same sequence but different structural condition information to RoseTTAFold, and used the linear combination of output logic values as the input for the next time step. The final ProteinGenerator generated protein sequences with different folding methods for the parent and child sequences. Researchers characterized 72 parent-child triplets through experiments, which were in a parent state when intact and in a child state when split. They selected four soluble monomer sequence families (MS1-MS4) for circular dichroism (CD) and nuclear magnetic resonance (NMR) testing. The experimental results indicate that all parent and child sequences of MS1-MS4 have good folding properties and undergo large-scale structural rearrangement during splitting, which is consistent with the design prediction.
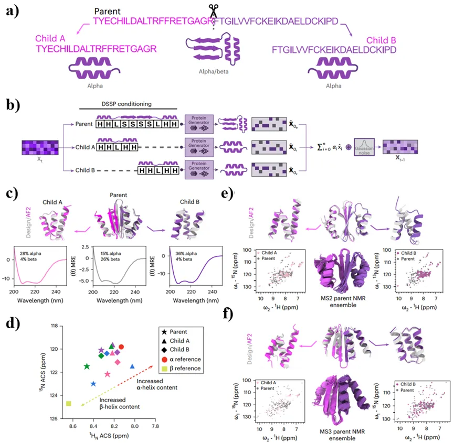 Figure 6. Design of an active peptide cage containing bee venom peptide. (a) (b) The principle and process of generating multi state protein sequences using ProteinGenerator; (c) The proportion of secondary structures in each state of MS1; (d) The chemical shifts corresponding to the parent and child structures in MS1-MS4; (e) (f) The structures of the parent and child sequences generated by MS1 and MS3, as well as the corresponding NMR results.
Figure 6. Design of an active peptide cage containing bee venom peptide. (a) (b) The principle and process of generating multi state protein sequences using ProteinGenerator; (c) The proportion of secondary structures in each state of MS1; (d) The chemical shifts corresponding to the parent and child structures in MS1-MS4; (e) (f) The structures of the parent and child sequences generated by MS1 and MS3, as well as the corresponding NMR results.
summary
The ProteinGenerator model generates protein sequences with expected structure and function by diffusing in sequence space and combining them with the three-dimensional structural information predicted by RoseTTAFold. Numerous experiments have shown that ProteinGenerator can handle a variety of complex protein design tasks, such as generating rare amino acids, repetitive proteins, designing bioactive peptide cages, and multi state proteins. Its efficiency, flexibility, and scalability make the ProteinGenerator model have broad application prospects in the field of protein design. This provides new tools and methods for drug design, biosensor development, and directed evolution experiments.
By editorRead more on
- Gan & Lee Pharmaceuticals’ new PROTAC drug GLR2037 tablets have been approved for clinical trials to enter the field of prostate cancer treatment March 3, 2026
- AideaPharmaceuticals plans to raise no more than 1.277 billion yuan through a private placement to focus on the global clinical development of innovative HIV drugs March 3, 2026
- Giant Exits! Its Star Business Acquired March 3, 2026
- Focusing on cardiovascular and cerebrovascular diseases! OpenMediLead Medical Intelligence Dual Engines Launch Internal Testing, Connecting Drug Development and Clinical Diagnosis in a Closed Loop March 3, 2026
- Innovent Biologics Announces Approval of New Indication for BTK Inhibitor “Pitubrutinib” in China March 3, 2026
your submission has already been received.
OK
Subscribe
Please enter a valid Email address!
Submit
The most relevant industry news & insight will be sent to you every two weeks.



