Chinese scientists release transcriptome map of single-cell space in the cerebellar cortex across species
September 29, 2024
Source: drugdu
 441
441
The cerebellum plays a crucial role in motor and cognitive functions, with region specific functional connections between its cortex and various sensory motor and associative regions of the neocortex. However, compared to the cerebral cortex, the various regions of the cerebellar cortex have relatively uniform cellular structures and typical local circuits, which makes the complexity of how cellular tissue supports cerebellar connections and functions an important issue.
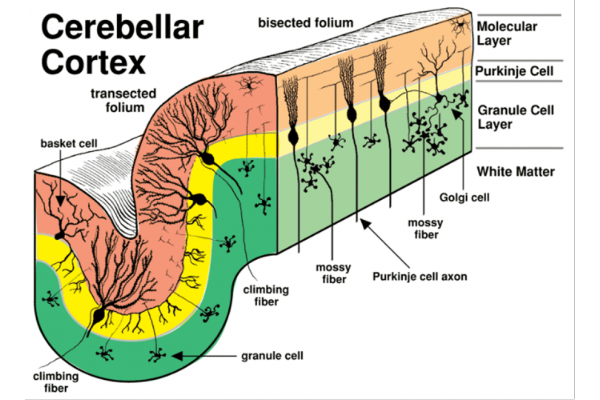 Figure 1: Composition of neurons in the cerebellar cortex
Figure 1: Composition of neurons in the cerebellar cortex
(Image reference from: https://vanat.ahc.umn.edu/neurLab6/Circuits.html )
In recent years, the development of single-cell omics has driven multiple studies on the composition of cerebellar cells, but previous research on the molecular and functional diversity of cerebellar cortical cells has mainly been based on the mouse cerebellum. The cerebellum of primates has significantly expanded during evolution, with an increase in the number of neurons and more complex cognitive functions. Therefore, determining the existence of primate specific cell subtypes and their unique spatial organization forms is of great significance for studying primate specific cerebellar function. However, due to the difficulty in enriching most cell types in the cerebellum except for granulosa cells, which account for over 99% of the cerebellar neurons, comprehensive spatial gene expression profiling analysis of the entire primate cerebellum at single-cell resolution has always been challenging.
On September 27, Science Journal published a research paper entitled "Cross specifications single cell spatial translational atlases of the cerebellar core" online. The research results were completed by a research team of nearly 100 people from the Chinese Academy of Sciences Brain Science and Intelligent Technology Innovation Excellence Center (Institute of Neuroscience), Hangzhou Huada Institute of Life Sciences, Shanghai Brain Science and Brain like Research Center, Chinese Academy of Sciences Key Laboratory of Genetic Evolution and Animal Models, Lingang Laboratory, South China University of Technology, Tencent AI Lab and other units.

In this study, scientists used the single-cell spatial transcriptome technique Stereo seq invented by the Huada Institute of Life Sciences to analyze the cerebellum of mice, macaques, and macaques. This method is capable of mapping the spatial gene expression profile of the cerebellum with a high spatial resolution of approximately 500 nanometers and a large visible area covering the entire cerebellum. By classifying the cell types in the cerebellar cortex of these species, researchers revealed conserved cell types such as Purkinje cells, granulosa cells, and intermediate neurons in the molecular layer among the three species. By comparing the gene expression patterns of each cell type in mice and primates, it was found that most cell types are conserved across species, but the differentially expressed genes in each cell type are highly species dependent. By plotting the spatial distribution of various cell types, researchers discovered the distribution characteristics of Golgi neurons specific to primates.
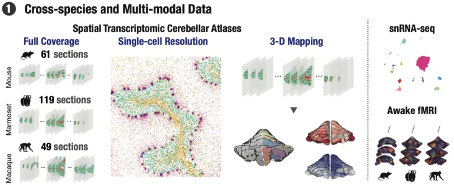
Figure 2: Cross species multimodal map of the cerebellum
Of particular note, researchers discovered a primate specific Purkinje cell subtype (GRID2 high Purkinje) in Stereo seq data, which was validated through RNAscope experiments and human single-cell transcriptome data. Further research also revealed significant differences in the expression of Long Term Inhibition (LTD) - related genes induced by GRID2 overexpressing cells, as well as differences in expression patterns with other glutamate receptor subunits.
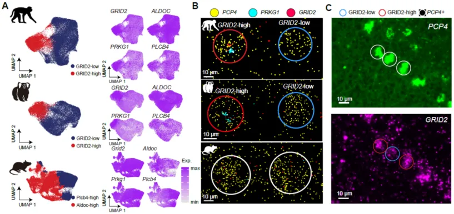 Figure 3: Primate specific Purkinje neuron subtypes
Figure 3: Primate specific Purkinje neuron subtypes
By collecting and analyzing resting state fMRI data from conscious mice, marmosets, and publicly available macaques, researchers discovered a functional connectivity gradient within the cerebellar cortex, known as the "cerebellar functional gradient". These gradients demonstrate the three-dimensional spatial patterns of connectivity between different cerebellar regions. By implementing three-dimensional reconstruction and gradient analysis of spatial transcriptomes, researchers further discovered that functional gradient 1 and transcriptome gradient 1 showed significant before and after differences in all three species, and the spatial pattern similarity between macaques and marmosets was higher than their similarity with mice. In addition, through the validation of cross species prediction models, a close relationship between gene expression and cerebellar functional connectivity was discovered, and genes related to functional gradients were identified. The expression patterns and enriched cell types of these genes vary among different species, further emphasizing the molecular and functional differences between species.
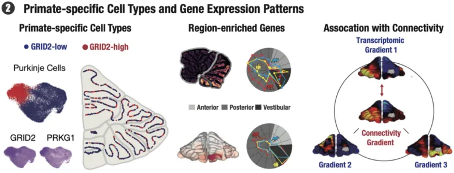 Figure 4: Important findings of this study
Figure 4: Important findings of this study
In summary, this study constructed spatiotemporal single-cell maps of the cerebellar cortex in macaques, marmosets, and mice, depicting primate specific Purkinje neuronal subtypes, evolutionarily conserved neuronal spatial patterns, and regional preferences for gene expression. The study also established a close relationship between transcriptome spatial heterogeneity and cerebellar functional connectivity. These data provide comprehensive resources for future research in this field, revealing the molecular and cellular diversity of the cerebellar cortex during its evolution, and contributing to a deeper understanding of the evolution and function of the cerebellum.
The research results have been published online, and the relevant data has been publicly submitted to the Shenzhen National Gene Bank, which can be accessed at CBMSTA( https://db.cngb.org/stomics/cbmsta/ )Interactively view and download database analysis.
This research achievement is of great significance for promoting the development of cerebellar research, providing important theoretical and experimental basis for further exploring the role of the cerebellum in the nervous system and the mechanisms of related diseases.
Liu Cirong, Sun Yidi, Liu Zhiyong and Shen Zhiming, researchers of the Brain Intelligence Center of Excellence (Institute of Neuroscience) of the Chinese Academy of Sciences, Liu Shiping and Liu Longqi, researchers of the Institute of Life Sciences of Hangzhou Huada, and Professor Chris I. De Zeeuw, Department of Neuroscience, Erasmus University, Rotterdam, are co corresponding authors of this paper. Hao Shijie, Associate Researcher at Hangzhou Huada Institute of Life Sciences, Huang Zhi and Wu Yan, Joint PhD Students, Zhu Xiaojia and Yang Qianqian, Joint PhD Students at the Brain Intelligence Excellence Center, Zhan Yafeng and Dong Yu, Postdoctoral Researchers, Li Chao, Director of Single Cell Platform, and Liu He'an, Research Assistant, are the co first authors of this paper.
Read more on
- Gan & Lee Pharmaceuticals’ new PROTAC drug GLR2037 tablets have been approved for clinical trials to enter the field of prostate cancer treatment March 3, 2026
- AideaPharmaceuticals plans to raise no more than 1.277 billion yuan through a private placement to focus on the global clinical development of innovative HIV drugs March 3, 2026
- Giant Exits! Its Star Business Acquired March 3, 2026
- Focusing on cardiovascular and cerebrovascular diseases! OpenMediLead Medical Intelligence Dual Engines Launch Internal Testing, Connecting Drug Development and Clinical Diagnosis in a Closed Loop March 3, 2026
- Innovent Biologics Announces Approval of New Indication for BTK Inhibitor “Pitubrutinib” in China March 3, 2026
your submission has already been received.
OK
Subscribe
Please enter a valid Email address!
Submit
The most relevant industry news & insight will be sent to you every two weeks.



